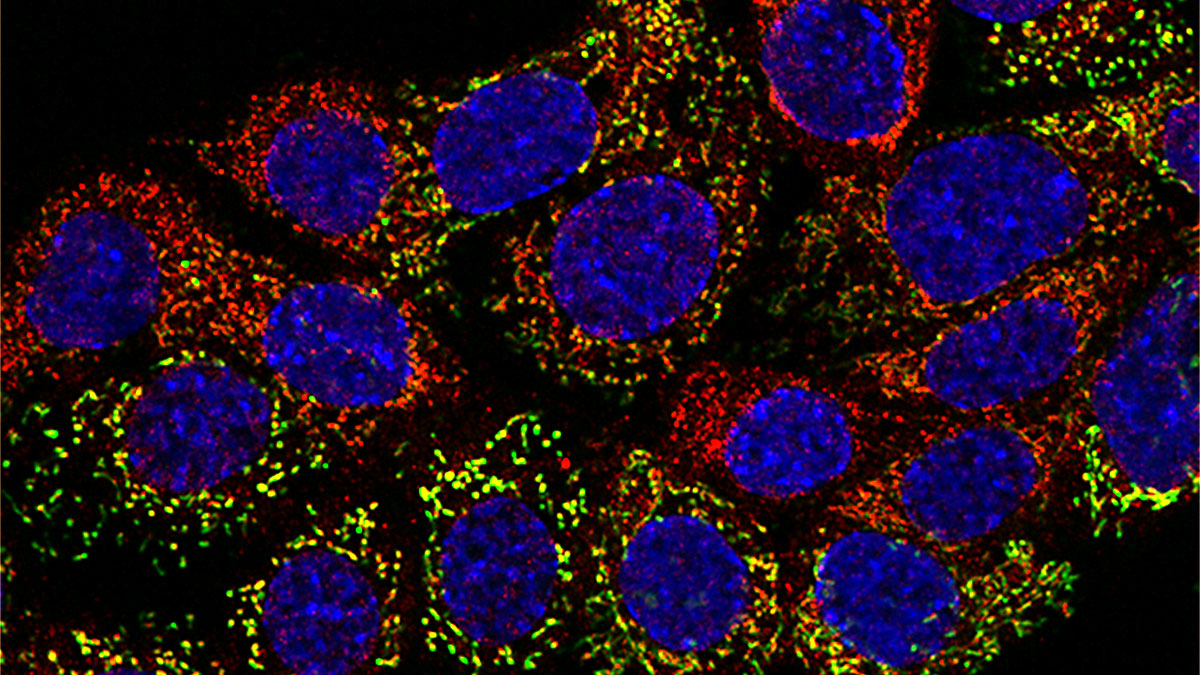
Turning Off Translation of Mitochondrial Genes
A paper1 recently published in Science explored the fascinating question of how the mitochondrial genome affects cellular function. The research work was carried out at the University Medical Center of Gottingen, led by Prof. Peter Rehling and Luis Daniel Cruz-Zaragoza (currently Professor at the Department of Biology, Université de Sherbrooke). Through a multi-institutional collaboration, the team developed a method to silence the translation of specific mitochondrial mRNAs selectively and used it to gain a deeper understanding of how mitochondria regulate translation.
Human mitochondria have their own small circular genome that encodes genes for essential proteins needed for oxidative phosphorylation. However, they are also involved in many other cellular functions, and most of the genes for those other functions are contained in the nuclear genome. The interactions of the two genomes are one of the most interesting open questions in biology today. Dissecting the translation of mitochondrial mRNAs has been challenging because standard methods (e.g., CRISPR) do not work inside mitochondria.
To overcome this challenge, the scientific team used synthetic peptide-morpholino chimeras to inhibit translation of specific mitochondrial genes. These synthetic molecules combine the phosphorodiamidate morpholino oligonucleotides directed against a specific mRNA with a mitochondrial targeting signal (or presequence). The chimeric molecules entered the mitochondria and blocked translation. By inhibiting the synthesis of each protein under different conditions, the researchers could ascertain the particular cellular response upon depletion.
With this study, the team identified proteins involved in the biogenesis and activity of the oxidative phosphorylation complexes. They also gained insights into how the cell deals with the loss of a key protein. Ultimately, the method can be applied to numerous other studies in mitochondrial biology.
Conversation with Dr. Cruz-Zaragoza
MitoWorld: Polymorpholino oligonucleotides are typically used as antisense oligonucleotides. Why did you decide to apply them for targeting the expression of mitochondrially encoded genes?
LDCZ: Close to 99% of proteins in the human mitochondria are encoded in the nuclear genome, translated in the cytosol, and imported into the mitochondria. Several genetic approaches are available to study and understand the function of this group of proteins, including CRISPR-Cas9, siRNA, epigenomic expression, and genomic integration. Those methods require the use of nucleic acids. Although transfecting nucleic acids into a cell is relatively straightforward, the same doesn’t apply to the mitochondria.
Mitochondria have two membranes: the outer mitochondrial membrane (OMM) and the inner mitochondrial membrane (IMM). The electron transport chain (ETC) is localized on the IMM. The activity of ETC generates a potential difference across the mitochondrial membrane, known as the mitochondrial membrane potential, making the IMM’s luminal side negatively charged. Therefore, it is no surprise that the introduction of nucleic acids into the mitochondria is highly inefficient, hindering efforts to apply genetic engineering to mitochondrial gene expression. Remarkably, researchers have developed several solutions based on proteins, such as base editors and mito-zinc finger protein nucleases.
We expected that non-charged oligonucleotides, such as phosphorodiamidate morpholino oligonucleotides, should be better imported into mitochondria. To target the morpholinos (MO) to the mitochondria, we initially fused the MO to a protein carrier via click chemistry, generating protein-MO chimeras. We applied this initial approach in isolated mitochondria.2
However, it did not escape our attention that a more versatile method was required for use in living cells. Therefore, building on our previous work, we optimized the chimera stability and efficiency by replacing the protein component with a mitochondrial targeting signal peptide.1
MitoWorld: What experiments do you have planned to extend this work?
LDCZ: It was thrilling to develop this expression-silencing project. For our recently published work, I had the opportunity to work with several talented graduate students. In addition, the collaborators who participated in our study are remarkable, combining expertise in proteomics (Bettina Warscheid’s group), microscopy (Stefan Jakob’s group), and transcriptomic data analysis (Michael Lidschreiber’s group). So, exciting work is in the making at University Medical Center of Gottingen.
I recently established my lab at the Université de Sherbrooke in Canada. As part of our research program, we will address the functional aspects of mitochondrial RNA life using the peptide-MO chimeras. We will follow an RNA-centered approach and examine how it correlates with the traditional protein-centered one. Luckily, Quebec, and Sherbrooke in particular, has an excellent group of scientists working in RNA biology as part of the RiboClub and the DNA to RNA Initiatives. Exciting times ahead!
MitoWorld: Were you able to link the loss of a specific protein to any human disease?
LDCZ: In our work, we explored the cellular response to challenges associated with the inhibition of subunits encoded in the mitochondrial genome. We have not yet made a direct comparison with mtDNA human disease models. However, it is an attractive problem that we, and other labs, will follow up on. One exciting aspect of our approach is the possibility of revealing and understanding, at the cellular level, the initial stages of defects in mitochondrial gene expression. In the field, researchers traditionally study patient-derived cells that have undergone long-term adaptation. However, silencing mitochondrial transcripts recapitulates the initial chronic depletion of proteins produced in the mitochondria, allowing us to examine how the cell responds and adapts to the challenge. In fact, one can study the onset of the disease in specialized cells, such as cardiomyocytes and hepatocytes. Therefore, this could be extended to other cell types, such as neurons and oocytes. This could contribute to the development of better therapies for mitochondrial diseases.
MitoWorld: Do you see any clinical or diagnostic applications for this methodology?
LDCZ: That is a great question. We can now tune the expression of specific genes encoded in mtDNA. This would allow for controlling the assembly rate of proteins with different genetic origins (nuclear and mtDNA) that form the respiratory complexes, which could be an avenue for treating disorders where the balance is lost. However, some aspects must be addressed before. The most important one is to show that the silencing approach is working in animal models.
MitoWorld: What drew you to mitochondria in the first place?
LDCZ: I have always been intrigued by the eukaryotic cell compartments. During my doctoral studies in Ralf Erdmann’s group at Ruhr-Universität Bochum, I investigated the peroxisome, focusing on its biogenesis mechanisms and function. I enjoyed it very much as part of the Marie Curie Initial Training Network PERFUME (PERoxisome, FUnction, MEtabolism). While attending conferences about protein targeting, I saw how exciting the research in mitochondria was. After finishing my PhD, I joined Prof. Rehling’s group to study mitochondrial protein import. Later, I became interested in working on mitochondrial gene expression. Combining my scientific interests in protein import and gene expression was central to devising the strategy of importing protein-MO chimeras to regulate mitochondrial gene expression.
References
1Cruz-Zaragoza LD, Dahal D, Koschel M, Boshnakovska A, Zheenbekova A, Yilmaz M, Morgenstern M, Dohrke JN, Bender J, Valpadashi A, Henningfeld KA, Oeljeklaus S, Kremer LS, Breuer M, Urbach O, Dennerlein S, Lidschreiber M, Jakobs S, Warscheid B, Rehling P (2025) Silencing mitochondrial gene expression in living cells. Science DOI: 10.1126/science.adr3498.
2Cruz-Zaragoza LD, Dennerlein S, Linden A, Yousefi R, Lavdovskaia E, Aich A, Falk RR, Gomkale R, Schöndorf T, Bohnsack MT, Ricarda Richter-Dennerlein R, Henning Urlaub H, Peter Rehling P (2021) An in vitro system to silence mitochondrial gene expression. Cell 184: 5824–5837.