New Pathway Mapping Program Links Complex II to Purine Biosynthesis
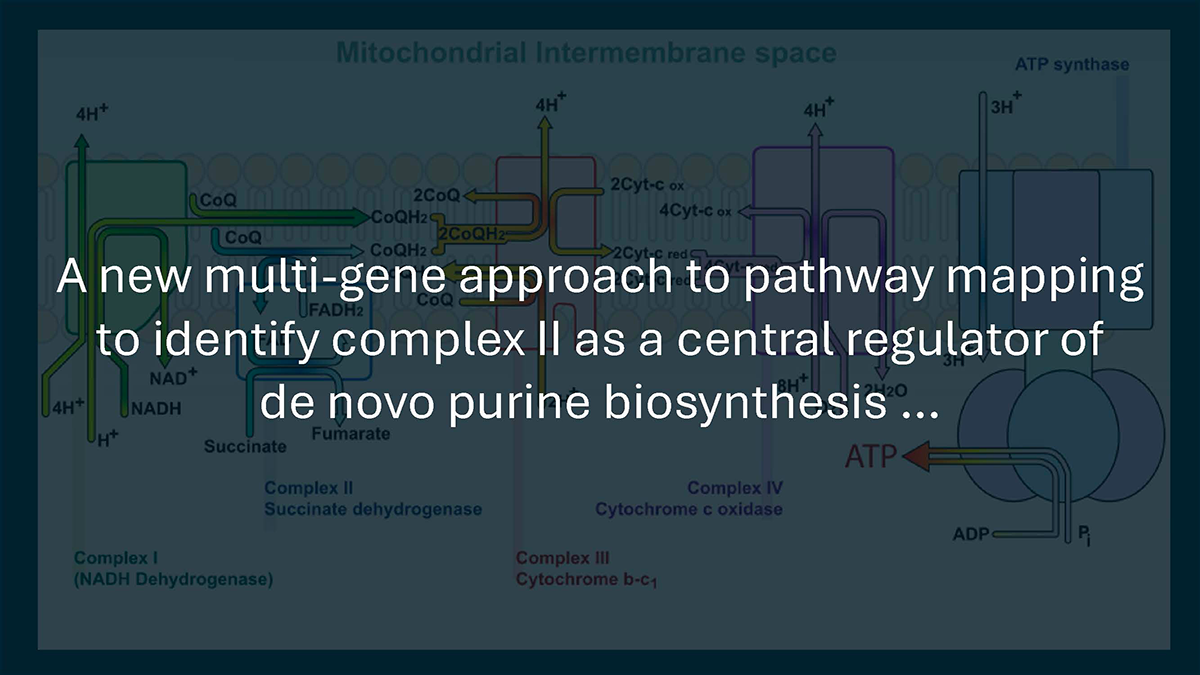
In a recent paper in Nature Metabolism, an international research group, used a new multi-gene approach to pathway mapping to identify complex II as a central regulator of de novo purine biosynthesis and a promising therapeutic target for acute myeloid leukemia.
Understanding genetic pathways is important in biology and medicine. However, complex diseases, such as cancer, might involve interactions between different pathways that are not immediately obvious. While much work has been done in understanding how individual genes interact, less is known about how multi-gene pathways interact.
Now a team, led by Matthew Hirschey and Kris Wood of Duke University and Alexandre Puissant of the Saint-Louis Research Institute/Hospital, INSERM in Paris, have developed a tool that can look for interactions between pathways. Specifically, they developed novel computational methods to examine genetic pathways involved in acute myeloid leukemia.
Interestingly, they discovered an unexpected link between complex II and purine metabolism with glutamate as a key intermediate. This observation implicates complex II as a potential therapeutic target. AML patients with higher complex II expression have worse survival rates.
These findings clearly demonstrate the power of data-driven tools for identifying critical interactions between genetic pathways. They also show the value this approach for finding possible new therapeutic strategies.
A conversation with Dr. Hirschey
MitoWorld: Congratulations on taking this next step in systems biology. Have you received any feedback from others in the field about it?
Hirschey: Thank you. The response has been very positive, particularly from colleagues working on cancer metabolism. Many have noted that our pathway-level approach fills a gap; while gene-gene coessentiality has been powerful, the emergent properties of pathways working together were being missed. Several groups have already started using our web tool at datadrivenhypothesis.org to explore their own systems of interest.
MitoWorld: Your work yielded surprising connections for glutamate and complex II. Do you have any other systems in mind to look at with this new tool?
Hirschey: Absolutely. Our analysis revealed Complex II has additional unique pathway connections that other ETC complexes don’t share. Beyond nucleotide metabolism, we saw strong links to amino acid pathways, particularly aspartate and glutamine metabolism. We’re interested in exploring how other TCA cycle enzymes might have similar “moonlighting” functions. Recent work from other groups on OGDH and fumarate hydratase suggests this is a rich area.
MitoWorld: You note several reasons that complex II is a possible therapeutic target in AML. Do you have any plans to follow up this line of research?
Hirschey: Yes, we’re actively pursuing this. Our data showing that Complex II inhibition sensitizes AML cells to venetoclax are particularly exciting given venetoclax resistance is a major clinical problem. We’re working on in vivo pharmacological studies and exploring synergistic strategies.
MitoWorld: Complex II may indeed be a target for AML and some other cancer. Can you speculate on whether it might be for a broader range of cancers?
Hirschey: Our pan-cancer analysis suggests caution here. High SDHB expression significantly increases mortality risk in only two cancer types, with AML being one. This contrasts sharply with pheochromocytoma and renal cancers, where Complex II acts as a tumor suppressor. So this isn’t a “one-size-fits-all” target. That said, our data point to broader vulnerability in hematolymphoid malignancies, where B-ALL, DLBCL, and anaplastic large cell lymphoma all showed Complex II dependency.
MitoWorld: In your discussion, you mention the increasing number of non-canonical functions of the mitochondria that have been found in recent years. Can you speculate on others that might be awaiting discovery?
Hirschey: I think we’re just scratching the surface. The TCA cycle has traditionally been viewed as an energy-producing pathway, but it’s clearly a biosynthetic hub with sensing functions. The finding that both FH deficiency and SDH inhibition suppress purine synthesis through metabolite accumulation suggests mitochondria may act as metabolic sensors, detecting imbalances through substrate buildup. I suspect we’ll find more examples where TCA intermediates serve as signaling molecules regulating distant pathways.
MitoWorld: We are obviously interested in mitochondria, but do you have any plans to examine pathways not involved with mitochondria?
Hirschey: Our pathway coessentiality tool is agnostic and can examine any gene set provided. We’ve already looked at transcription factor targets and cell-type signatures beyond metabolism. I’m particularly interested in how metabolic pathways connect to epigenetic regulation, given the known links between TCA metabolites, such as succinate and α-ketoglutarate to chromatin-modifying enzymes. The tool is publicly available, so we hope others will explore non-mitochondrial systems as well.
Reference
Stewart AE, Zachman DK, Castellano-Escuder P, Kelly LM, Zolyomi B, Aiduk MDI, Delaney CD, Lock IC, Bosc C, Bradley J, Killarney ST, Stuart JD, Grimsrud PA, Ilkayeva OR, Newgard CB, Chandel NS, Puissant A, Wood KC, Hirschey MD (2025) Pathway coessentiality mapping reveals complex II is required for de novo purine biosynthesis in acute myeloid leukaemia. Nat Metab 7: 2474–2488 (2025). https://doi.org/10.1038/s42255-025-01410-x.